R10.3 and ‘R10m’ Data Release (2020-02-07)
New R10.3 Pore
Summary stats:
- Platform ONT GridION
- Flowcells 2
- Reads 1.16 M
- Bases 4.64 Gbp
- Read Length N50 25,163 bp
- Mean Quality 11.5
Download links:
- Merged FASTQ Basecalls (4.8 GB,
78497b92a6b8fde2e0f5d2291c707949).
Prototype R10 pore with methylation-aware basecalling model
Prototype R10 pore data basecalled with Guppy 3.2.4+d9ed22f and model template_r10_450bps_hac_meth.
Summary stats:
- Platform ONT GridION
- Flowcells 2
- Reads 6.74 M
- Bases 30.6 Gbp
- Read Length N50 8,871 bp
- Mean Quality 9.5
Download links:
- Merged FASTQ Basecalls (32 GB,
eed7625452dc5a9b6f6313c9e7200fa2).
R10 Native “3 Peaks” Data Release (2019-05-24)
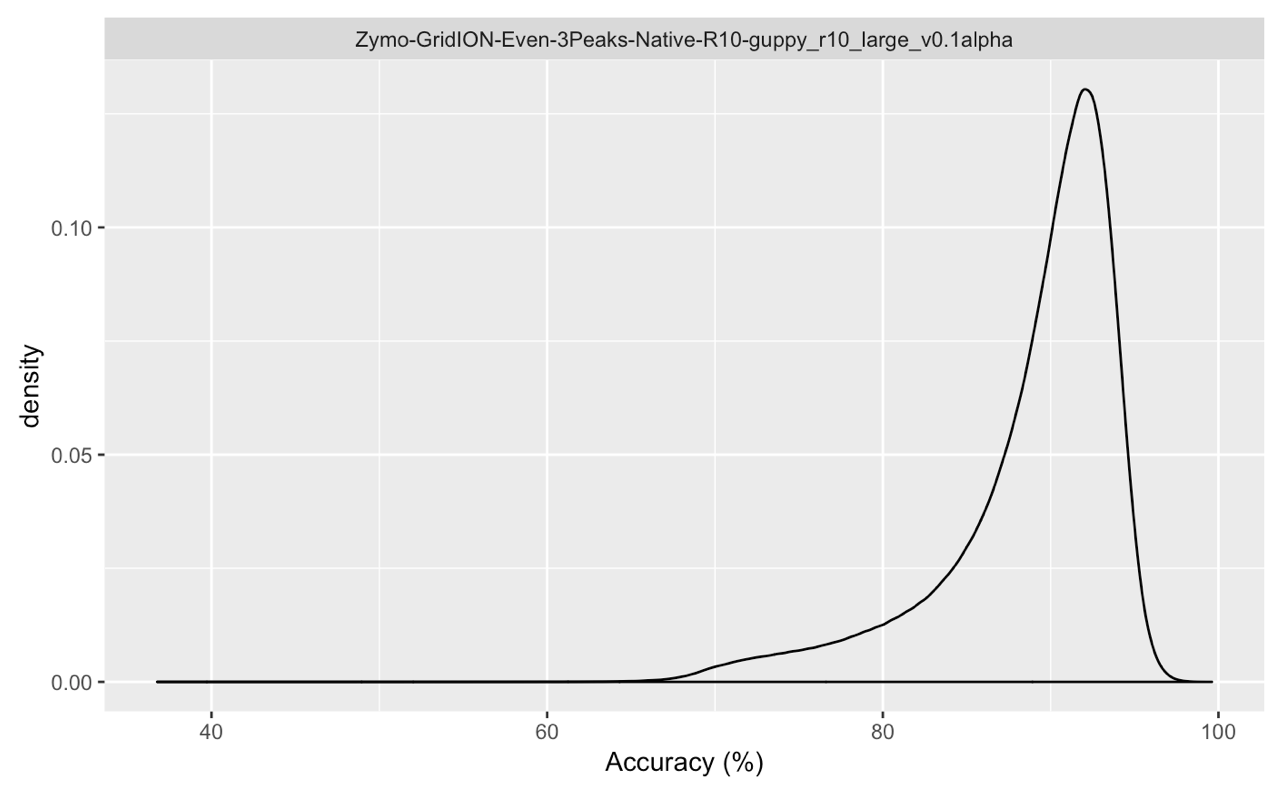
Summary stats:
- Reads 6.74 M
- Bases 30.9 Gbp
- Read Length N50 8893 bp
- Mean Quality 9.6
Singal files were basecalled with Guppy 2.3.8 and the basecalling model guppy_r10_large_v0.1alpha.
Download links:
- FAST5 Signal (375 GB)
- FASTQ Basecalls (33 GB,
f2a9c2a64f6104462a36622b53d8d424)
Please note this is a preliminary analysis of R10 data with alpha release models and a pre-release pore type and is subject to change!
R10 PCR Data Release (2019-02-28)

Summary stats:
- Reads 4.24 M
- Bases 16.6 Gbp
- Read length N50 4620 bp
- Median Quality 8.0
Signal files were basecalled using Guppy 2.3.1+1b9405b using a pre-release basecalling model R10_flipflop_model.json.
Download links (courtesy of CLIMB):
Please note this is a preliminary analysis of R10 data with alpha release models and a pre-release pore type and is subject to change!
R9.4 ~ R10 Comparison
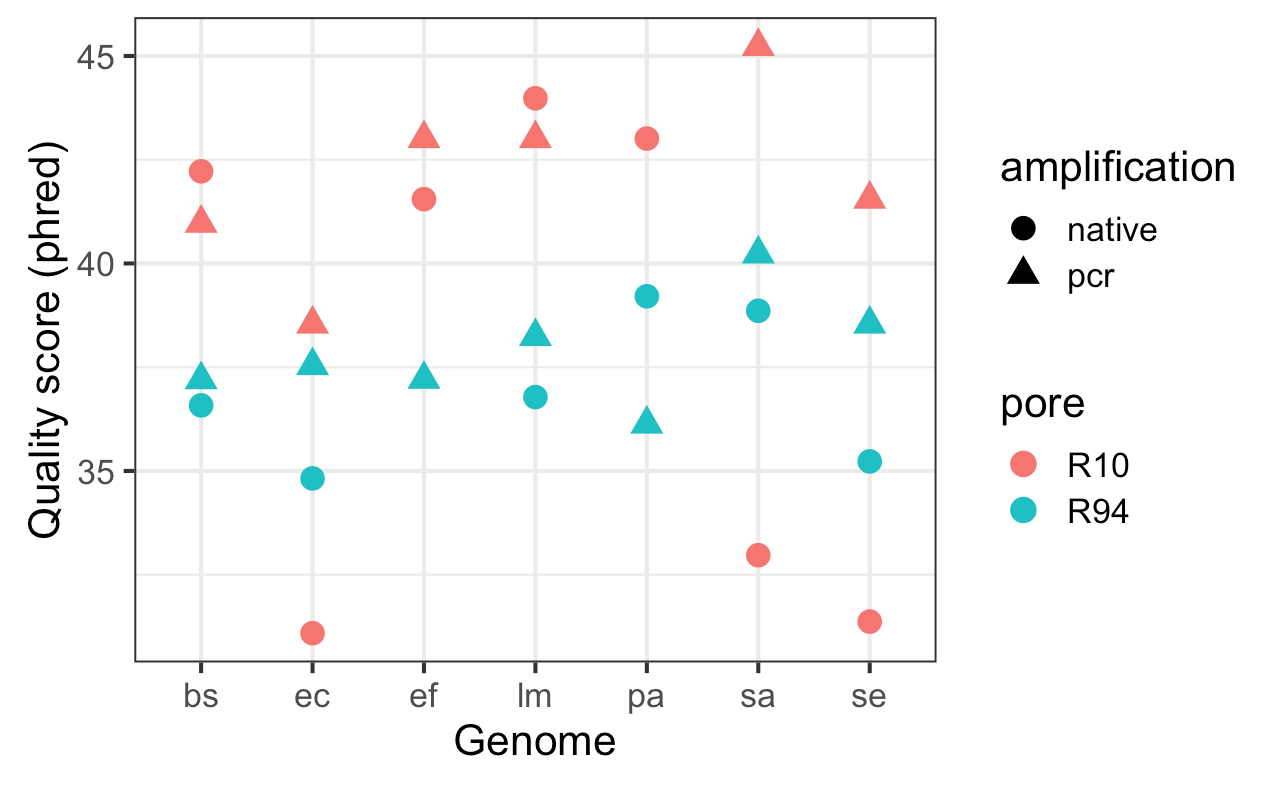
Reads from the Zymo mock community runs were binned by taxa using the Illumina draft assembly from our paper. Bins were downsampled to 200x and an assembly for each genome was built with wtdbg2 (v2.4) with the -x ont parameter.
After wtdbg2-poa consensus, assemblies were polished with four rounds of racon and one round of medaka (R10 -m medaka_r10_v0.1alpha.hdf, R9.4 -m r941_flip213).
Per-genome accuracy was measured with Jared Simpson’s FASTMER script.
Acknowledgements
We thank Rosemary Dokos, Chris Wright, Jon Pugh and Jayne Wallace from Oxford Nanopore Technologies for their help and assistance with preparation of these datasets.